
CIBM announces the co-organisation of two challenges at the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI2021) to be held in Strasbourg, France from 27 September to 1 October.
Diffusion-Simulated Connectivity Challenge (DiSCo), MICCAI 2021 COMPUTATIONAL DIFFUSION MRI
The methodological development in the mapping of the brain structural connectome from diffusion MRI has raised many hopes in the neuroscientific community. Quantifying the structural connectivity between brain regions is fundamental for studying brain anatomy and its function. The reliability of the structural connectome is therefore of paramount importance.
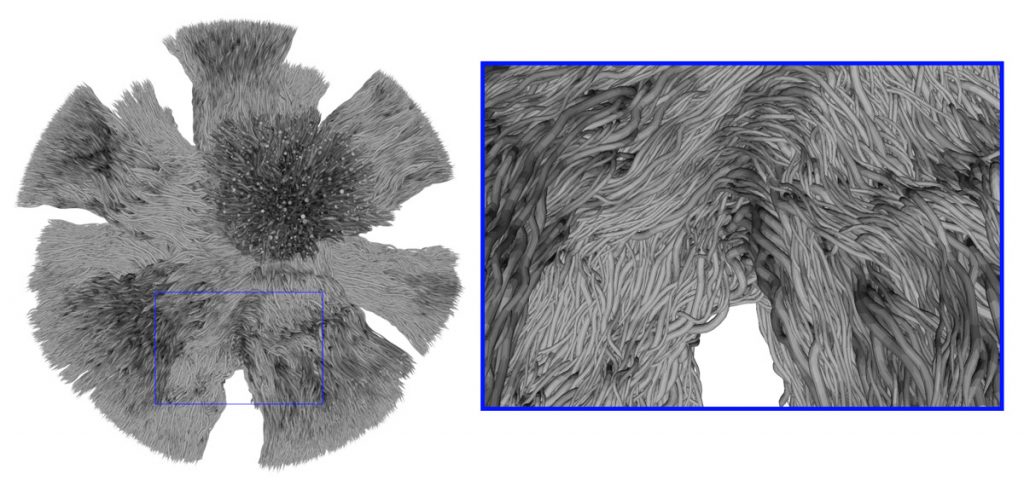
The main goal of the Diffusion-Simulated Connectivity (DiSCo) Challenge is to evaluate the performance of quantitative connectivity pipelines on numerical phantoms with known ground truth connectivity. The synthetic signal was obtained using Monte-Carlo simulations of spins dynamics in a substrate composed of a large collection of synthetic tubular fibres, interconnecting distant regions. This is the first time this technique was used to create diffusion MRI phantoms of such size and complexity. The simulated images capture microscopic properties of the white matter tissue, while also having desirable macroscopic properties resembling the anatomy.
Ideally, quantitative connectivity estimation for pairs of regions with few interconnecting synthetic fibres should be weaker than for pairs of areas with several interconnecting synthetic fibres. An evaluation and ranking in this sense could prompt the improvement of tractography methods for quantitative structural connectivity estimation in clinical or neuroscientific settings, where we expect to find connectivity estimates directly related to the number of interconnecting axons. Overall, the challenge will provide unique datasets and analysis to foster the development of tractography and connectivity estimation methods using the rich information available from the complex microstructural organization of the white matter.
The DiSCo Challenge is organized by Dr. Gabriel Girard (CIBM SP CHUV-EPFL Section), Jonathan Rafael-Patino (EPFL), Raphaël Truffet (Univ Rennes, Inria, CNRS, Inserm), Dr. Marco Pizzolato (Technical University of Denmark), Dr. Emmanuel Caruyer (Univ Rennes, Inria, CNRS, Inserm) and Prof. Jean-Philippe Thiran (EPFL, CIBM SP CHUV-EPFL Section).
Important dates:
- Release of the validation and test datasets: June 1st, 2021
- Registration deadline: September 1st, 2021
- Submission deadline: September 13th, 2021

Fetal Brain Tissue Annotation and Segmentation Challenge (FeTA), MICCAI 2021
The Fetal Brain Tissue Annotation and Segmentation Challenge (FeTA) is a multi-class image segmentation challenge organized as part of MICCAI 2021. The goal of FeTA is to compare automatic multi-class segmentation methods for the segmentation of developing human brain tissues. The challenge provides manually annotated, super-resolution reconstructed MRI data of human fetal brains which will be used for training and testing automated multi-class image segmentation algorithms.
The full challenge design can be found here: Fetal Brain Tissue Annotation and Segmentation Challenge | Zenodo
The FeTA Challenge is organized by Kelly Payette (University of Zurich & University Children’s Hospital Zurich), Priscille de Dumast (University of Lausanne), Andras Jakab, (University of Zurich & University Children’s Hospital Zurich), Meritxell Bach Cuadra (University of Lausanne & CIBM SP CHUV-UNIL Section), Roxane Licandro (TU Wien & Medical University of Vienna), Hongwei (Bran) Li (Technical University of Munich), Bjoern Menze (University of Zurich), Lana Vasung (Harvard Medical School)
Data Release
The data for the FeTA Challenge has been released! Click here for details: FeTA Dataset
Important Dates
- Data Release: May 3, 2021
- Docker Submission Deadline: July 26, 2021
- Algorithm Description Deadline: August 5, 2021
- Notification of Presentations to top teams: End of August/Beginning of September
