SOFTWARE
Electroencephalography (EEG)
Cartool
Cartool is an EEG analysis software entirely programmed in C++ and doesn’t rely on any other dependency to run.
The project started in 1996 at the Functional Brain Mapping Lab (FBMLab), University of Geneva Switzerland and completely developed by Denis Brunet, technical staff scientist at the CIBM EEG HUG-UNIGE Clinical and Translational Neuroimaging Section.
STEN
STEN (Statistical Toolbox for Electrical Neuroimaging) is an open source software toolbox based on Python and R that can be used to compute statistics on several measures of electro- and magneto-encephalographic (EEG and MEG) signals.
Signal Processing (SP)
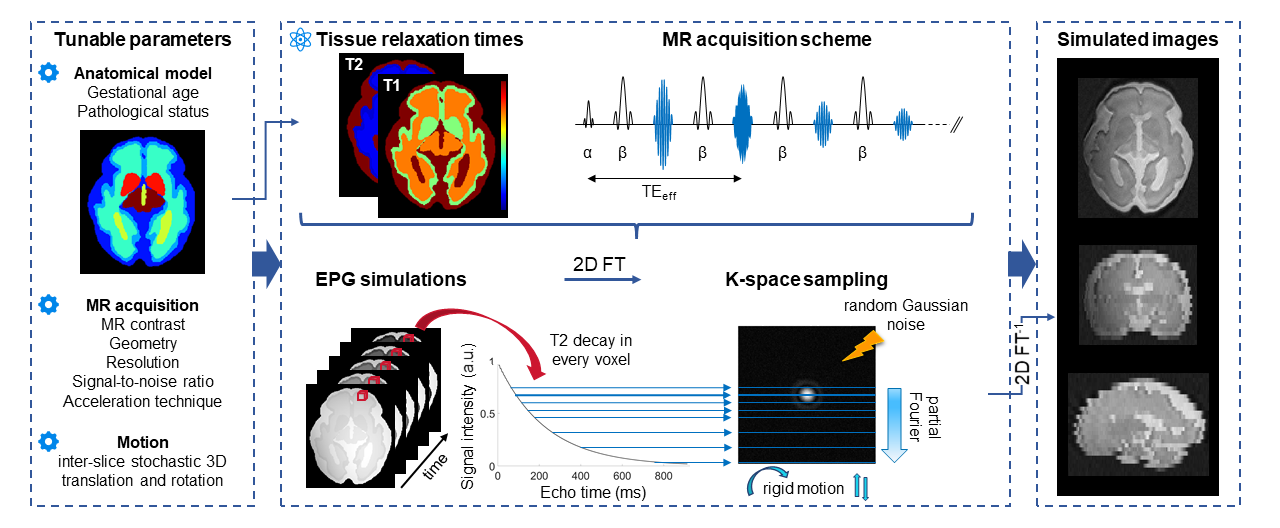
FaBiAN
FaBiAN is the first Fetal Brain magnetic resonance Acquisition Numerical phantom that simulates realistic T2-weighted magnetic resonance images (MRI) of the developing fetal brain throughout gestation. With this open-source simulation framework implemented in MATLAB (MathWorks, R2019a), we aim at providing the community with a unified and controlled environment for the evaluation of various post-processing techniques dedicated to improving the analysis of fetal brain MRI for accurate diagnosis. The synthetic, yet realistic fetal brain MR images generated using FaBiAN have also proven to be very useful in supplementing scarce clinical datasets to support data augmentation and domain adaptation strategies.
MIALSRTK
The Medical Image Analysis Laboratory Super-Resolution ToolKit (MIALSRTK) consists of a set of C++ image processing tools necessary to perform motion-robust super-resolution fetal MRI reconstruction. This toolkit, supported by the Swiss National Science Foundation (grant SNSF-141283), includes all algorithms and methods for brain extraction, intensity standardization, motion estimation and super-resolution developed during my PhD. It uses the CMake build system and depends on the open-source image processing Insight ToolKit (ITK) library, the command line parser TCLAP library and OpenMP for multi-threading.
GlobalBioIm Library
GlobalBioIm Library is a unifying Matlab library for imaging inverse problem developped by members of the CIBM Mathematical Imaging EPFL section.
When being confronted with a new inverse problem, the common experience is that one has to reimplement (if not reinvent) the wheel (=forward model + optimization algorithm), which is very time consuming and also acts as a deterrent for engaging in new developments. This Matlab library GlobalBioIm aims at simplifying this process by decomposing the workflow onto smaller modules, including many reusable ones since several aspects such as regularization and the injection of prior knowledge are rather generic. It also capitalizes on the strong commonalities between the various image formation models that can be exploited to obtain fast, streamlined implementations.
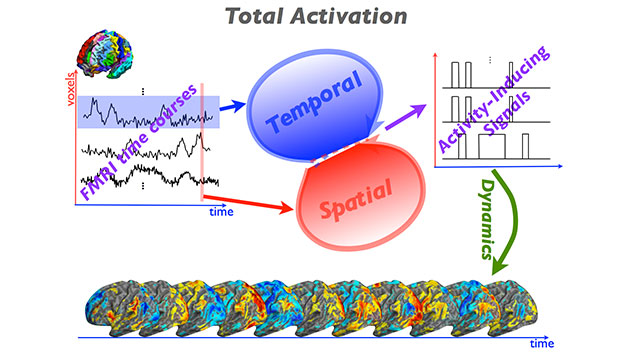
iCAPs Toolbox
The iCAPs Toolbox contains multiples libraries to provide a window into dynamics of functional magnetic resonance imaging (fMRI) and reveal the organizational principles of brain function. Using state-of-the-art sparsity-pursuing deconvolution, termed “Total Activation” (TA), we extract innovation-driven co-activation patterns (iCAPs) from resting-state fMRI. The iCAPs’ maps are spatially overlapping and their sustained-activity signals temporally overlapping. Decomposing fMRI data using iCAPs reveals the rich spatiotemporal structure of functional components that dynamically assemble known resting-state/intrinsic networks. The temporal overlap between iCAPs is substantial and is consistent with their behavioral profiles.
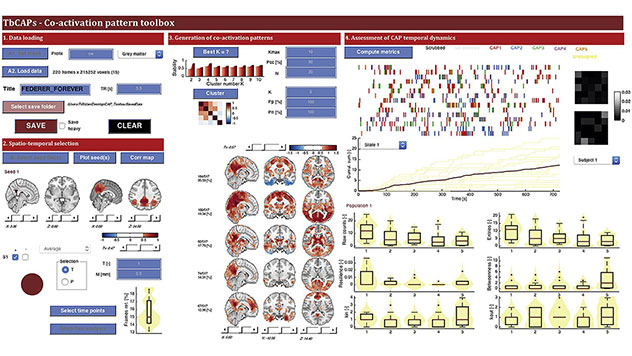
TbCAPs
The Matlab Toolbox for Co-Activation Patterns (TbCAPs) provides a frame-wise analytical approach that disentangles the different functional brain networks interacting with a user-defined seed region. While promising applications in various clinical settings have been demonstrated, there is not yet any centralised, publicly accessible resource to facilitate the deployment of the technique, which is one of the objectives of TbCAPs.
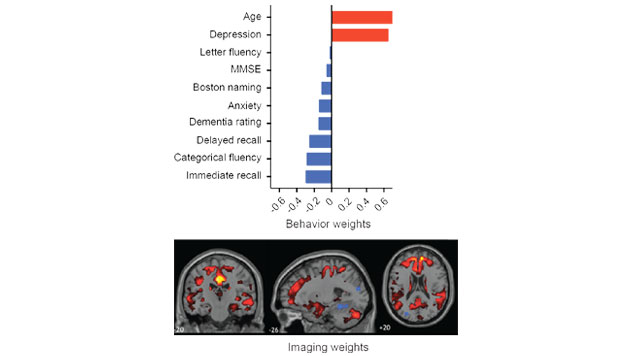
myPLS
PLS correlation is a powerful approach for linking data from two modalities. The Matlab myPLS Toolbox is a user-friendly set of command line tools that incorporate a multitude of options for the application of PLS correlation to different types of neuroimaging data.